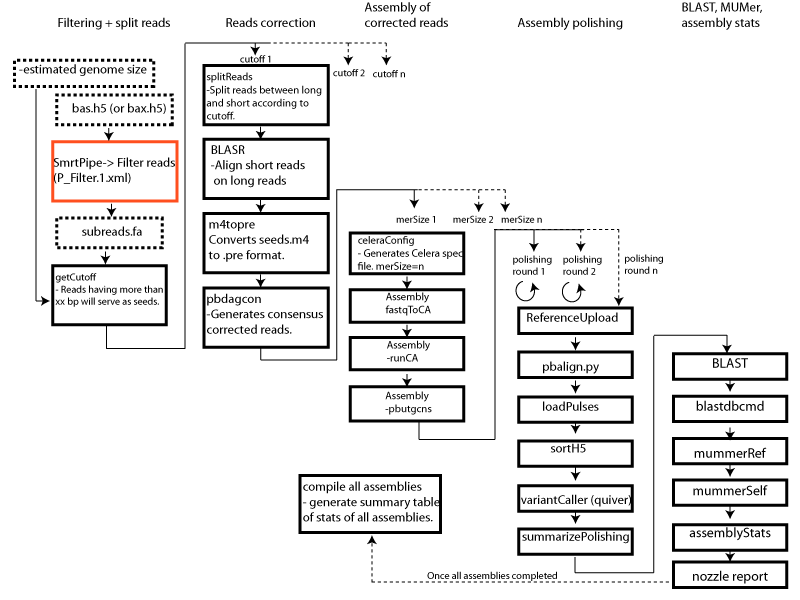
PacBio HGAP3 assembly pipeline
I’ve recently been working on implementing the PacBio HGAP genome assembly pipeline on our pipeline infrastructure here at the McGill University – Genome Quebec Innovation Centre (https://bitbucket.org/mugqic) . Anyone who has done analysis on PacBio data is certainly familiar with PacBio’s Smrtanalysis (smrtpipe) software/pipeline suite. While this pipeline performs well, it is, from a bioinformatician point of view, a little cumbersome to use. All those xml protocol files are not super intuitive. Plus you don’t get to see what’s happening in details when you run from these xml protocol files. To shed some light on that black box, I ran an HGAP assembly with smrtpipe and looked into the logs to see what was really happening in there. Having in hands every single commands ran by the HGAP xml protocol, I could then write a pipeline that could give me flexibility (i.e. generate multiple assemblies with different long reads cutoffs and use various kmer sizes for the corrected reads assembly). So here, without going into details on what the pipeline does (you can go have a look at the code from our repo), I thought I could share some details on this. Here is a figure summarizing the pipeline’s steps.